Abstract
Introduction High-grade B-cell lymphoma (HGBCL) with MYC and BCL2 and/or BCL6 rearrangement was established as a diagnostic entity in the 2017 WHO-HAEM4 classification. Updated classifications (WHO-HAEM5 and International Consensus Classification [ICC]) have further refined this entity to include only tumors with MYC and BCL2 rearrangement (irrespective of BCL6 status) (HGBCL-BCL2) - albeit with the creation of a new provisional ICC entity for tumors with MYC and BCL6 rearrangement (HGBCL-BCL6). Diagnosis of HGBCL-BCL2 requires fluorescence in situ hybridization (FISH) testing in all tumors with diffuse large B-cell (DLBCL) or high-grade morphology, most commonly performed using FISH break-apart probes (BAPs). Typical MYC BAPs include a red (R) and green (G) probe which hybridize to regions centromeric and telomeric to MYC (8q24.21), respectively. When the MYC locus is intact, 2 fused (F) signals are observed, while separation of a fused signal is indicative of MYC rearrangement (MYC-R) (Figure 1A). Although MYC-R typically produces a balanced BAP pattern (equal R and G signals, e.g. 1F1R1G), a subset of tumors display unbalanced patterns which can be categorized into 3 groups, 1) increased R or G signal (gain-R/G, e.g. 1F2R1G, 1F1R2G), loss of R signal (LR, e.g. 1F1G), or loss of G signal (LG, e.g. 1F1R). As LR and LG patterns indicate loss of genetic material, these patterns have produced uncertainty about whether a MYC-R is present. Understanding the significance of LR and LG patterns is needed as the diagnosis of HGBCL-BCL2 carries important clinical implications, such as these patients commonly receiving intensified treatment.
Methods 531 MYC-R tumors with DLBCL or high-grade morphology were identified from the BC Cancer Lymphoid Cancer database. FISH was performed with commercially available BAPs. MYC-R partner and breakpoint location were detected from whole genome or capture sequencing using Manta and GRIDSS2. MYC protein expression was determined by immunohistochemistry (IHC; ≥40% cutoff), and MYC mRNA expression was determined by NanoString.
Results Among all MYC-R tumors, 69% of MYC BAP patterns were balanced, 17% gain-R/G, 10% LG, and 4% LR. BCL2 and BCL6 FISH was available for 95% (505/531) of biopsies, 60% of which were HGBCL-BCL2, 10% were HGBCL-BCL6, and 30% were MYC single hit.
Among HGBCL-BCL2 tumors, the proportion of each MYC BAP pattern was similar to the proportions observed across all MYC-R tumors, with 65% balanced, 22% gain-R/G, 11% LG, and 3% LR. To assess the significance of the LG and LR patterns in HGBCL-BCL2, MYC protein and MYC mRNA expression levels were compared to HGBCL-BCL2 tumors with balanced patterns, as well as 241 MYC break-apart negative GCB DLBCL. There was no significant difference in MYC mRNA expression levels (LR: p = 0.79; LG: p = 0.30; t-test) and proportions of MYC IHC positivity (LR: p = 1; LG: p = 0.40; Fisher's exact) in LR or LG groups relative to tumors with a balanced pattern, while both groups had higher MYC mRNA (LR: p = 0.02; LG: p < 0.0001) and IHC positivity (LR: p = 0.0001; LG: < 0.0001) compared to GCB DLBCL.
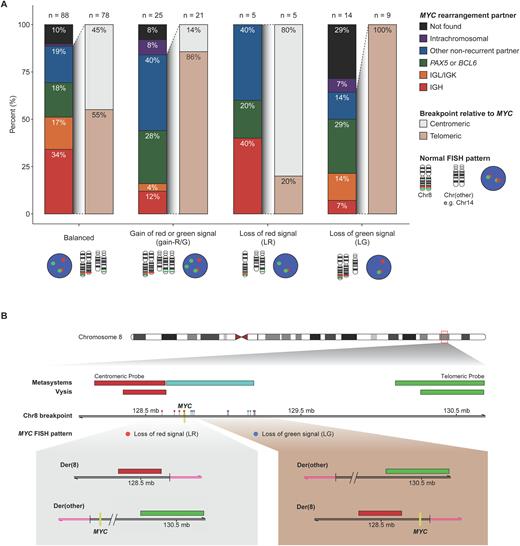
Sequencing was available for 44% (132/302) of HGBCL-BCL2 tumors. A translocation partner was detected for 88% (78/88) of tumors with a balanced pattern, compared to 64% (9/14) of tumors with an LG pattern (p = 0.66; Fisher's exact), and 100% (5/5) with an LR pattern (Figure 1A). Among tumors with an identified translocation partner, the breakpoint was centromeric to MYC in 80% (4/5) of tumors with an LR pattern, while 100% (9/9) of tumors with an LG pattern had a breakpoint telomeric to MYC (Figure 1A and B). For a centromeric break, the resulting rearrangement places MYC on the derivative chromosome containing the G signal, while for a telomeric break, MYC is placed on the derivative chromosome containing the R signal (Figure 1B). As such, the MYC gene was preserved in 93% (13/14) of tumors with an LR or LG pattern.
Conclusion An unbalanced LR or LG MYC FISH pattern was observed in 14% of HGBCL-BCL2. These tumors displayed similar MYC protein and MYC mRNA expression levels as HGBCL-BCL2 tumors with balanced MYC FISH patterns. While detection of a translocation in sequencing data can be limited by factors such as poor biopsy quality, a translocation partner was identified for the majority of tumors with LR and LG patterns, with all but 1 translocation preserving the MYC gene. These results support that LR and LG patterns should be interpreted as a positive MYC FISH result.
Disclosures
Craig:BeiGene: Honoraria; Bayer: Consultancy. Villa:Roche, AstraZeneca, Abbvie, Janssen, Kite/Gilead, BMS/Celgene, BeiGene, Kyowa Kirin: Consultancy, Honoraria; AstraZeneca, Roche: Research Funding. Gerrie:AbbVie: Honoraria, Research Funding; AstraZeneca: Honoraria, Research Funding; Janssen: Honoraria, Research Funding; Sandoz: Honoraria. Sehn:AbbVie, Acerta, Amgen, Apobiologix, AstraZeneca, BMS/Celgene, Gilead, Incyte, Janssen, Kite, Karyopharm, Lundbeck, Merck, Morphosys, Sandoz, Seattle Genetics, Servier, Takeda, TG Therapeutics, Verastem: Honoraria; Chugai: Consultancy, Honoraria; AbbVie, Acerta, Amgen, Apobiologix, AstraZeneca, BMS/Celgene, Debiopharm, Genmab, Gilead, Incyte, Janssen, Kite, Karyopharm, Lundbeck, Merck, Morphosys, Novartis, Sandoz, Seattle Genetics, Servier, Takeda, TG Therapeutics, Verastem: Consultancy; Teva, Roche/Genentech: Consultancy, Honoraria, Research Funding. Savage:Beigene and Regeneron: Membership on an entity's Board of Directors or advisory committees; BMS, Janssen, Kyowa, Merck, Novartis, and Seattle Genetics: Consultancy, Membership on an entity's Board of Directors or advisory committees. Steidl:AbbVie: Consultancy; Bayer: Consultancy; Bristol-Myers Squibb: Consultancy; Curis Inc: Consultancy; Epizyme: Research Funding; Roche: Consultancy; Seattle Genetics: Consultancy; Trillium Therapeutics: Research Funding. Scott:Incyte: Consultancy; AstraZeneca: Consultancy, Honoraria; Abbvie: Consultancy; Janssen: Consultancy, Research Funding; Roche: Research Funding; NanoString: Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal